Introduction
There is always a big debate between which language, R or Python, is the best for statistical data analysis and machine learning. Both languages have pros and cons, so why not understand both? I have a strong Python background, but figured I should learn R as well. R has ggplot2, which is an amazing visualization library that seems to outdo what matplotlib and Seaborn offer through Python. Unfortunately, there seems to be a lot of information about switching from R to Python but not the other way around. So I decided to just learn R through rebuilding a Python project I have already completed.
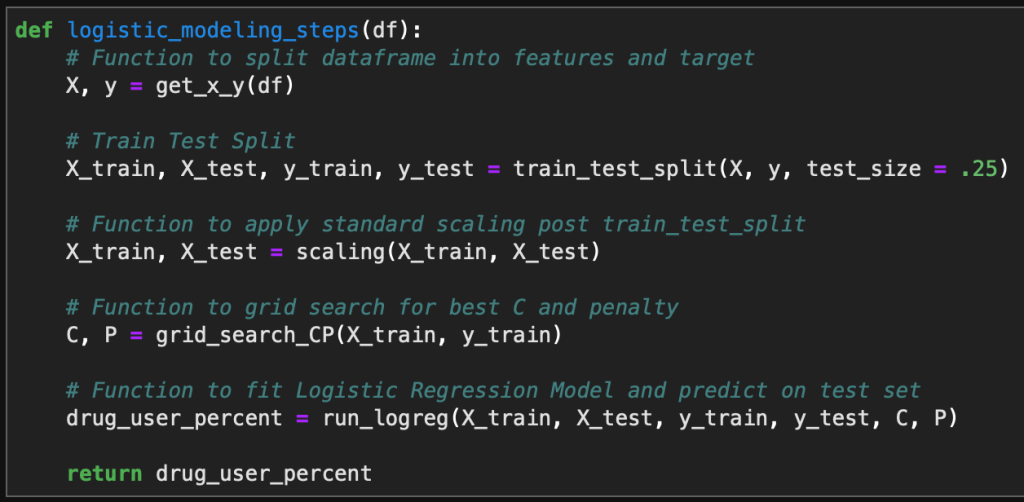
For reference, I built a spam classifier model in Python and documented the process in a previous blog. I wanted to rebuild this spam classifier model, in a very simplified form, in R. Doing this has helped me learn many useful skills in R that I will show in this blog.
Getting Started
To begin, I downloaded Python through Anaconda. I typically code with jupyter notebooks, which come with Anaconda. I’d like to set up R in this environment as well.
First things first, let’s download R. Then in order to install properly follow the PC or Mac steps found in this helpful blog by Rich Pauloo.
Coding with R
Installing Libraries
If you need to install any of the libraries I’ll use, then you can do this with the following line of code, just change the library name:
install.packages("caret")
Importing Libraries
I’ll be using the following libraries:
library(wordcloud)
library(RColorBrewer)
library(tm)
library(magrittr)
library(caret)
library(e1071)
library(SnowballC)
Reading in the data
I had the spam data in a csv file from my python project, but the data originally comes from the UCI Machine Learning Repository.
I read in the data with the following code:
df <- read.csv("smsspamcollection/spamham.csv")
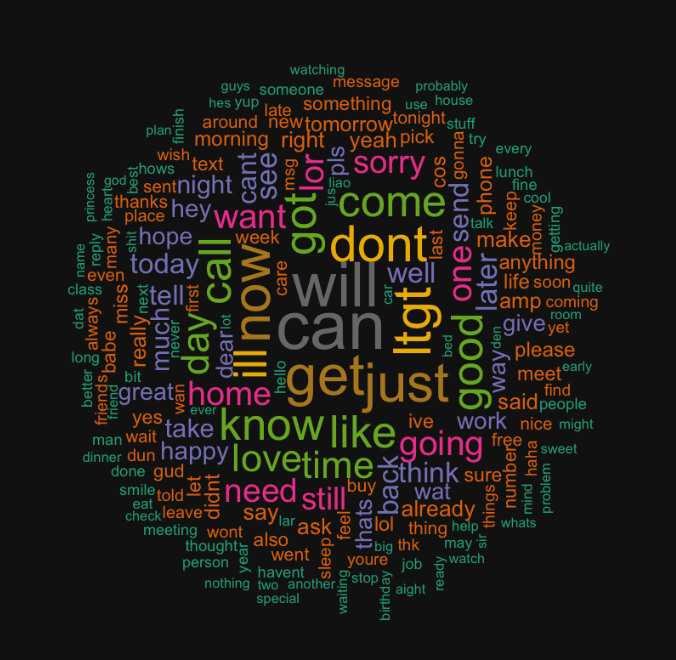
Data Visualizations through Wordcloud Library
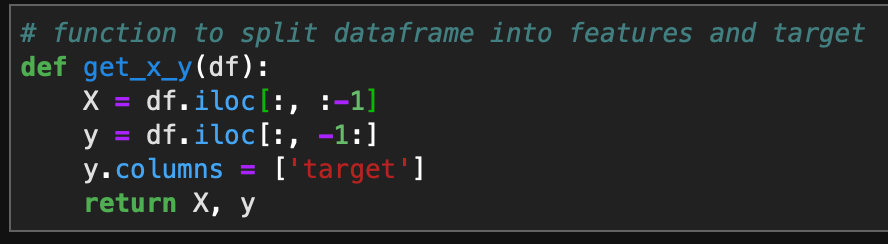
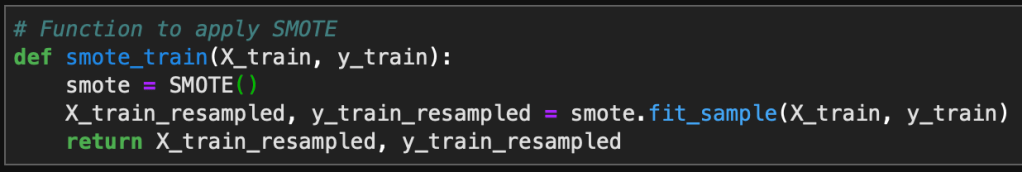
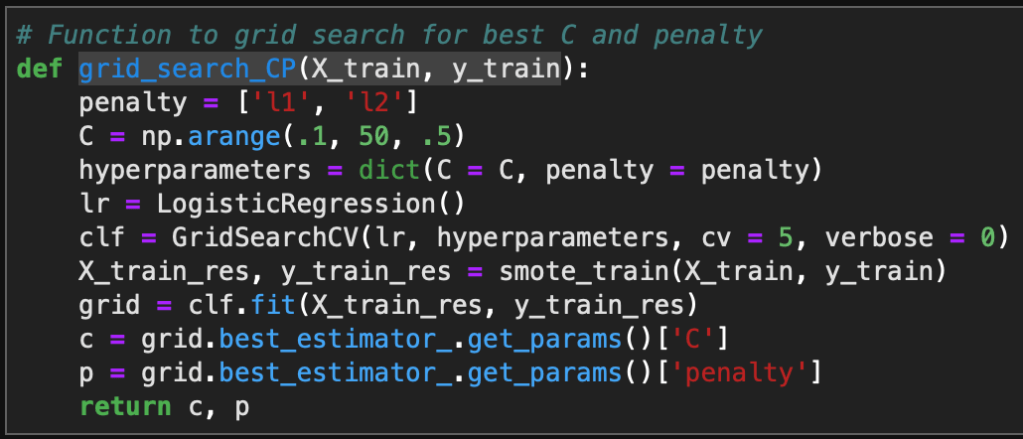
Now in order to visualize the text data, I separated the data into spam vs ham (i.e. not spam) and then created a word cloud for each group. This was accomplished with the following code:
# Split dataframe by ham and spam
spam_split <- split(df, df$label)
spam <- spam_split$spam
ham <- spam_split$ham
# Create a vector of just spam text data
spam_text <- spam$text
# Create a spam corpus
spam_docs <- Corpus(VectorSource(spam_text))
# Clean spam text with tm library
spam_docs <- spam_docs %>%
tm_map(removeNumbers) %>%
tm_map(removePunctuation) %>%
tm_map(stripWhitespace)
spam_docs <- tm_map(spam_docs, content_transformer(tolower))
spam_docs <- tm_map(spam_docs, removeWords, stopwords("english"))
# Create document spam term matrix
spam_dtm <- TermDocumentMatrix(spam_docs)
spam_matrix <- as.matrix(spam_dtm)
spam_words <- sort(rowSums(spam_matrix),decreasing=TRUE)
spam_df <- data.frame(word = names(spam_words),freq = spam_words)
# Create a vector of just ham text data
ham_text <- ham$text
# Create a ham corpus
ham_docs <- Corpus(VectorSource(ham_text))
# Clean ham text with tm library
ham_docs <- ham_docs %>%
tm_map(removeNumbers) %>%
tm_map(removePunctuation) %>%
tm_map(stripWhitespace)
ham_docs <- tm_map(ham_docs, content_transformer(tolower))
ham_docs <- tm_map(ham_docs, removeWords, stopwords("english"))
# Create document ham term matrix
ham_dtm <- TermDocumentMatrix(ham_docs)
ham_matrix <- as.matrix(ham_dtm)
ham_words <- sort(rowSums(ham_matrix),decreasing=TRUE)
ham_df <- data.frame(word = names(ham_words),freq = ham_words)
# Create spam wordcloud
wordcloud(words = spam_df$word, freq = spam_df$freq, min.freq = 1,
max.words = 200, random.order = FALSE, rot.per = 0.35,
colors = brewer.pal(8, "Dark2"))
# Create ham wordcloud
wordcloud(words = ham_df$word, freq = ham_df$freq, min.freq = 1,
max.words = 200, random.order = FALSE, rot.per = 0.35,
colors = brewer.pal(8, "Dark2"))
This code should give you two word clouds, spam and ham respectively, that look like the following:

Model Preprocessing
Now I need to do some preprocessing for the modeling. This includes creating a corpus and document term matrix, which is a sparse matrix containing all the words and the frequency of which they appear in each message. The preprocessing code also removes stop words (i.e. the, it, etc.), makes all words lowercase, finds the stem of the words, and removes punctuation and numbers.
df_corpus <- VCorpus(VectorSource(df$text))
df_dtm <- DocumentTermMatrix(df_corpus, control =
list(tolower = TRUE,
removeNumbers = TRUE,
stopwords = TRUE,
removePunctuation = TRUE,
stemming = TRUE))
I also need to train test split the data. This step is admittedly much less straightforward as compared to Python, but I was able to get it done.
# Calculate train test proportions
index <- floor(5572 * .8)
#Training & Test set
train <- df_dtm[1:index, ]
test <- df_dtm[index:5572, ]
#Training & Test Label
train_labels <- df[1:index, ]$label
test_labels <- df[index:5572, ]$label
Finally, the last step of preprocessing is converting the data into categorical data. The Naive Bayes model I am using in R requires this. This step is good practice and creating and implementing a function!
# Convert to categorical for naive bayes model
convert_values <- function(x) {
x <- ifelse(x > 0, "Yes", "No")
}
train <- apply(train, MARGIN = 2, convert_values)
test <- apply(test, MARGIN = 2, convert_values)
Modeling
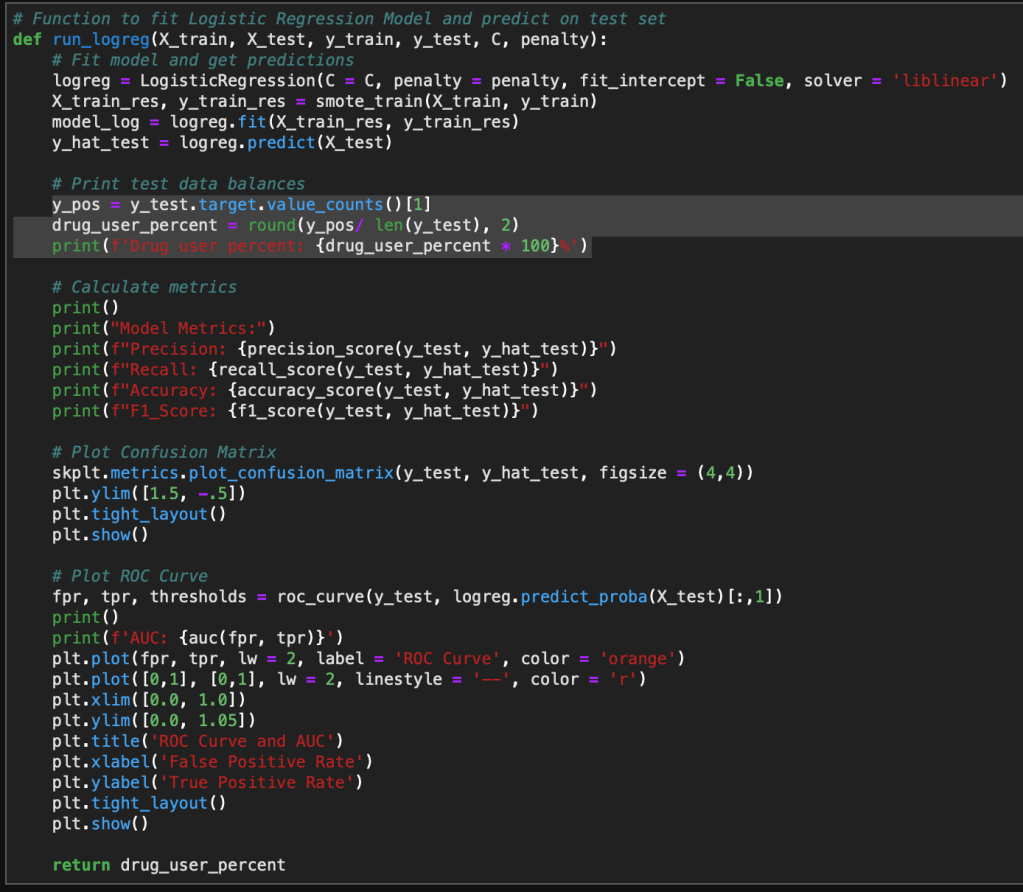
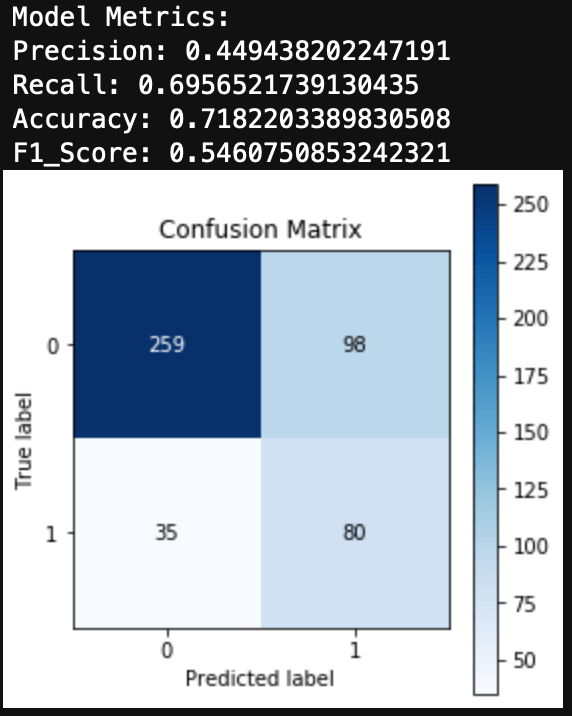
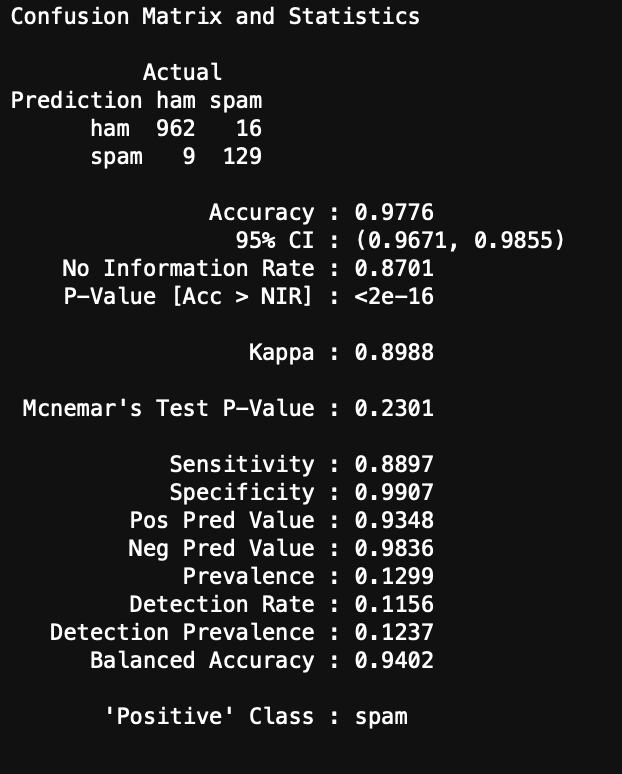
Now we can model and evaluate! The following code initiates the Naive Bayes model and computes a confusion matrix and accuracy of the Naive Bayes model on the test set of data.
#Create model from the training dataset
spam_classifier <- naiveBayes(train, train_labels)
#Make predictions on test set
y_hat_test <- predict(spam_classifier, test)
#Create confusion matrix
confusionMatrix(data = y_hat_test, reference = test_labels,
positive = "spam", dnn = c("Prediction", "Actual"))
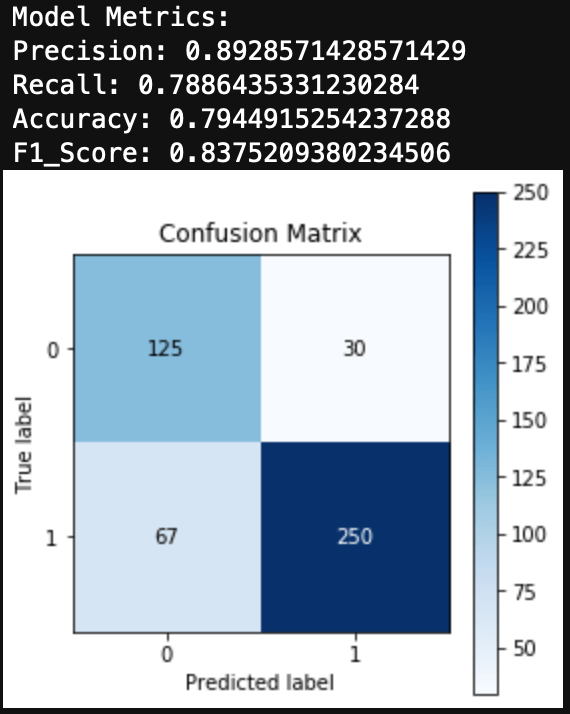
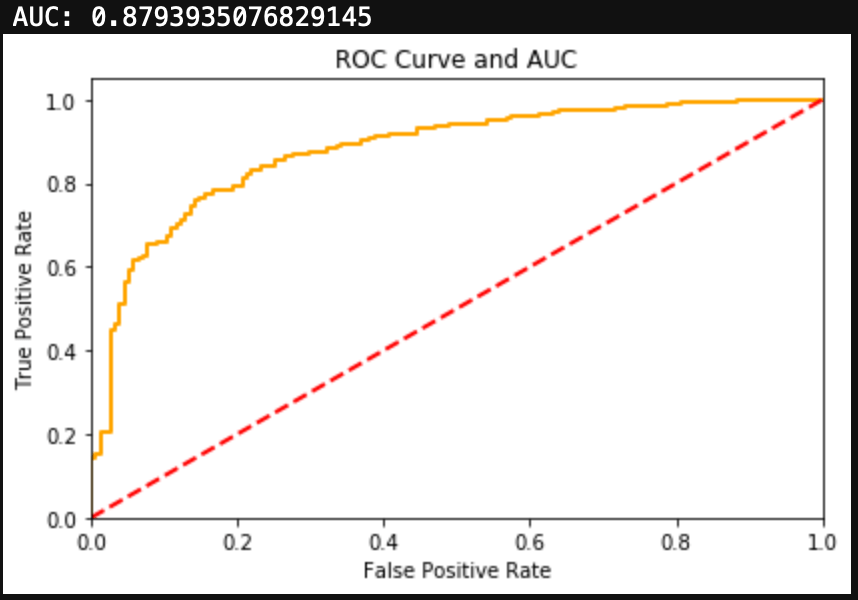
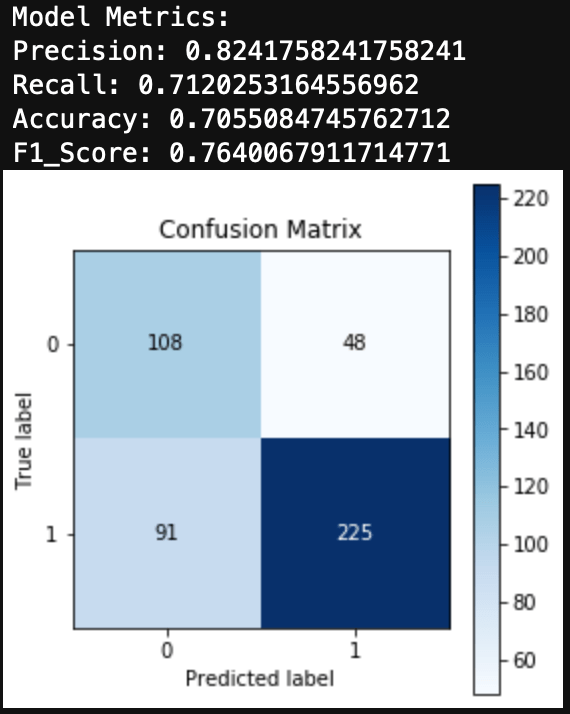
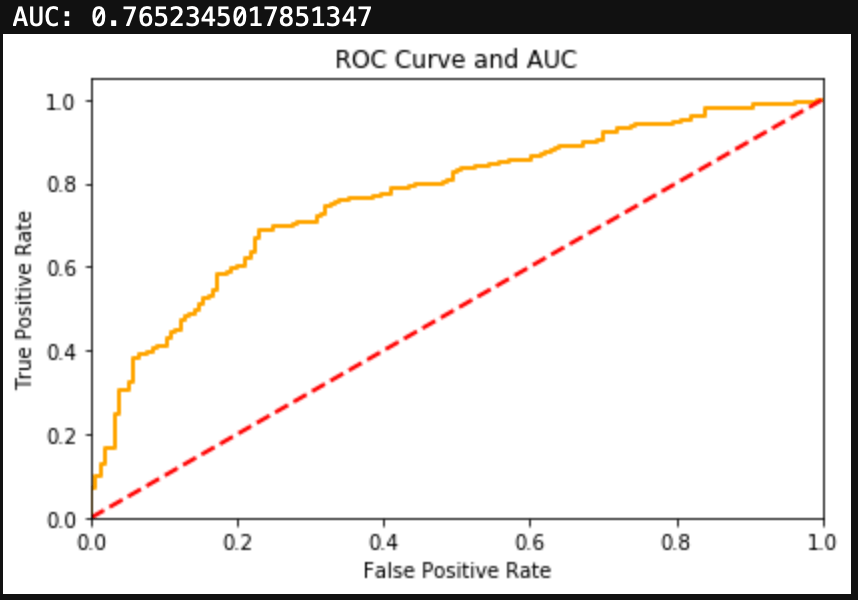
You can see the results here:
Conclusions
I was able to get a very simplified working model of my spam classifier! Although this isn’t as pretty as the model generated in Python from my previous blog, it still works and is a great success for learning R. If you are wanting to learn R from Python, I encourage you to practice R by recreating a Python project you have already created. This way you know what you want as data inputs and model outputs, and all you have to figure out is the R. Googling helps!